Abstract
The gene transcription of bacteria starts with a promoter sequence being recognized by a transcription factor found in the RNAP enzyme, this process is assisted through the conservation of nucleotides as well as other factors governing these intergenic regions. Faced with this, the coding of genetic information into physical aspects of the DNA such as enthalpy, stability, and base-pair stacking could suggest promoter activity as well as protrude differentiation of promoter and non-promoter data. In this work, a total of 3131 promoter sequences associated to six different sigma factors in the bacterium E. coli were converted into numeric attributes, a strong set of control sequences referring to a shuffled version of the original sequences as well as coding regions is provided. Then, the parameterized genetic information was normalized, exhaustively analyzed through statistical tests. The results suggest that strong signals in the promoter sequences match the binding site of transcription factor proteins, indicating that promoter activity is well represented by its conversion into physical attributes. Moreover, the features tested in this report conveyed significant variances between promoter and control data, enabling these features to be employed in bacterial promoter classification. The results produced here may aid in bacterial promoter recognition by providing a robust set of biological inferences.
Similar content being viewed by others
1 Introduction
The transcription of DNA into messenger RNA (mRNA) is a crucial step in the cellular gene expression. This process is finely regulated by a plethora of proteins that recognize specific promoters and operators located in the intergenic regions, in response to environmental changes [3, 4, 16, 20]. Promoter sequences are DNA segments located upstream of the transcription start site (TSS) or + 1, where the enzyme RNA polymerase (RNAP) attaches to carry out gene transcription. In bacteria, this interaction is only possible when an additional protein, a sigma (σ) factor, interacts with the RNAP. The primary role of σ factors is to redirect the RNAP to specific promoters, granting specificity to promoter recognition [16].
Promoters are usually conserved at the sequence level; for instance, the promoters associated with σ70 exhibit two consensual motifs around − 10 and − 35 (TATAAT, referred to as the Pribnow box and TTGACA, respectively), upstream from the TSS. However, the architecture of promoters is more diverse than expected. Some bacterial promoters, for example, present an overlapping of the promoter into the TSS, while others are distinguished by an absence of the − 35 region and an extension of the − 10 region (extended promoters) [14]. These variations, together with insertion and deletion of base pairs, as well as different mutations, may jeopardize any promoter identification task that mainly relies on the presence of specific nucleotides in a sequence window.
In spite of this, an in-silico analysis of promoters can include alternative approaches to investigate the DNA molecule and extract structural properties [28]. Some of these features have potential to enhance the sensitivity and specificity of the approach, including enthalpy, free-stability, base-pair stacking, stress-induced DNA duplex destabilization, curvature, and bending. Thus, the parametrization of DNA sequences into structural properties has shown promising results in capturing specific promoter DNA signals [33].
Recently, bioinformatics tools based on the structural architecture of promoters have been developed to aid in this field. Examples of these tools are PromPredict [27], BTSS Finder [31], and BacPP [8]. These approaches consider structural properties to distinguish promoters from other genomic regions. In general, these studies have shown that promoter sequences exhibit different structural properties when compared to their neighboring regions [2, 21, 33]. In this regard, the parametrization of the DNA into structural attributes is connected to gene expression variability [9, 27, 33]. Therefore, the computational re-coding of information stored in the DNA sequence might reveal a uniqueness between promoters in comparison to non-promoter data.
There are many features used to code genetic information into numeric attributes. However, energy-related features have been reported to yield differentiation unmatched by other forms. To this extent, we propose to employ enthalpy, free-stability, and base-pair stacking as explicit indicators of promoter activity, i.e., strong signals that match basal transcription factor binding sites, as well as capturing differences that turn promoter regions unparalleled.
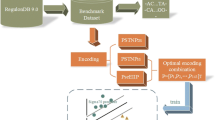
2 Datasets and methods
2.1 Promoter sequences
A total of 3131 promoter sequences associated with six different sigma factors of the bacterium Escherichia coli K-12 (Table 1) were downloaded from the database RegulonDB v. 9.0 [12]. We considered promoter sequences that are either experimentally observed or predicted in this research. σ19 was not considered in this analysis since no data was available in the database. RegulonDB also contains promoters recognized by more than one σ factor; in such cases, we only consider the sequences associated to the lower-molecular-weight entity. The promoter sequences available at RegulonDB contain 81 nucleotides (− 60 to+ 20). We decided to preserve this length due to bacterial promoters having extensively been reported to be found in this range [5, 19].
Additionally, we have also selected an experimentally validated collection of promoters regulated by σ54 in order to provide further validation to the stablished method. Pro54DB [18] holds up to 43 species with regulatory and transcriptional information, from this, we selected 15 Gram-negative bacteria to maintain the same structure of Gram-negative promoters previously identified [6].
2.2 Non-promoter sequences
We constructed a total of 3131 random sequences through a Python script (Supplementary Materials S1) that shuffles the original promoter sequences, maintaining the AT content and length size (81 nucleotides). Since each σ group of sequences is found in different proportions in RegulonDB; therefore, we opted to have the exact number of the promoter and non-promoter sequences. Moreover, coding sequences were also considered for enabling the genetic variance portrayed by promoters [33]. Transcriptome data were retrieved from published information on E. coli [15], and sequences with the same size of the promoter sequences were maintained. Therefore, coding sequences containing 81 nucleotides were randomly selected in the position downstream of + 20. The number of coding sequences was selected to match each σ group [17].
We opted to have two levels of control sequences: (i) a shuffled version of the promoter sequence, which has been achieved by the in-house script available in Supplementary Materials S1 and (ii) a same-sized (81 nucleotides) coding sequence aiming to capture the differences coding regions have when compared to promoters. Since each σ group is found in different proportions in RegulonDB, we opted to maintain the exact number of promoter and non-promoter sequences, granting validity to the experiment. The obtaining of downstream sequences followed the published transcriptome data [15], the maintenance of the same length of promoter and coding sequences enabled the distinction of transcription factor binding sites, which is only expected to occur within promoters.
2.3 Conversion into enthalpy, base-pair stacking and free-stability
We considered enthalpy, free-stability (average free energy), and base-pair stacking as structural attributes to codify promoter and non-promoter sequences into structural values. The nucleotide duplex values were achieved by DNA melting studies at 37 °C, and measured nearest-neighbor (NN) thermodynamics [1, 23, 29, 30]. The calculation considers NN, moving around the promoter sequence in windows containing two nucleotides and assigning a structural value for each position (the script used to this conversion is available in Supplementary Materials S2).
To this end, we used the following equations to calculate the DNA duplex free-stability (1), and enthalpy (2) scores of all observed dinucleotides in sliding overlapping windows (1-nucleotide step):
In addition, we evaluated the mean scores of the individual promoter and non-promoter sequences, where G is the free-stability variation and H is enthalpy. For each window, the free energy was calculated considering the above equations, obtained from the dinucleotide sequences from Allawi and SantaLucia [1] and SantaLucia and Hicks [29].
Furthermore, the base-pair stacking calculation (3) was performed according to the following equation [23]:
where A is the stacking energy, \(\Sigma a\) is the sum of all stacking values; and n is the number of dinucleotides [30].
2.4 Data normalization
The values in Table 2 show a difference in scale. For this purpose, we performed a normalization process to compare the four features together. The normalization technique employed is considered in Eq. (4), and transforms the data into values between 0 and 1:
where n is the result of the normalization process; v is the entropy, enthalpy, base-pair stacking, or free-stability value that is being normalized; min is the smallest value from the feature in Table 2, and max is the highest value from the feature in Table 2.
2.5 Data representation
A spreadsheet for each feature in each σ group was constructed. The means for each position of all promoters and non-promoters were calculated and analyzed.
2.6 Statistical evaluation
The nonparametric Spearman correlation test was employed to check the correlation of the three DNA features. Furthermore, the nonparametric Kruskal–Wallis test was performed in order to test the data variance and define if this variance is significant in order to differentiate the groups tested in this study. Both tests were done by using the R programming language through its stats package.
2.7 Sequence logo profiles
We built sequence logo profiles [7] to determine the sequence conservation around the consensual motifs for each promoter dataset.
3 Results and discussion
3.1 RNAP binding is explained by different levels of enthalpy, free-stability, and base-pair stacking
The average enthalpy, free-stability, and base-pair stacking values for each nucleotide position within the promoter were compared in order to have their importance highlighted in correctly representing promoter activity. From Fig. 1, it is observable that the three features showed a similar distribution pattern in all the promoters, with the exception of σ24 promoters. The entwined aspect found suggests the promoter activity is well represented and each one of the energy-related features might describe promoter activity. The employment of structural features has shown a satisfactory way to locate transcription factor (TF) protein binding sites [13] in a way sheer consensual motifs are not enough to represent promoter activity, indeed, Deyneko et al. [10] suggested that there is a feature conservation around promoter sequences.
In order to explain the super positioning observed in Fig. 1, a correlation analysis was performed. Since the data do not follow a normal distribution, the Spearman correlation test was chosen. From this analysis, the strongest correlation was found between enthalpy and base-pair stacking by presenting a Spearman’s Ro of 0.806, whereas enthalpy and stability presented a medium correlation (Spearman’s Ro = 0.53); and stability and base-pair stacking, showed a moderate correlation, by presenting Spearman = 0.478. In addition, σ24 promoters exhibited a considerable amount of noise in their structural feature profile and, unlike other σ factor regulated sequences, did not exhibit any perceivable overlapping between the features. In this particular case, around 80% of the σ24 promoters listed in RegulonDB were determined by prediction instead of being confirmed by either biological experiments or human inference.
Secondly, we suggest that RNAP binding sites by σ factors are indicated by variations in the trajectories represented by the lines in Fig. 1. Except for σ24, all promoters presented noticeable peaks. Promoter sequences regulated by σ28, σ32, σ38, and σ70 had strong signals observed in their − 10 vicinity. This transcription factor binding site was found to be the most conserved in E. coli promoters [19]. Additionally, Fig. 1 also indicates variations in position − 35 in σ28, σ32, and σ70. This position is another binding site for transcription factor proteins in bacterial promoters. Finally, the consensual motifs of σ54 (− 12 and − 24) were also well represented by Fig. 1.
Finally, two families of promoters were identified: (i) σ24, σ28, σ32, σ38, and σ70, which have presented strong signals in either − 10 or − 35 and; (ii) σ54, whose consensual region was found in − 12 and − 24. The promoters from the first family should, in theory, retain conservation in two TF binding sites. However, the results suggested that σ28 might be distinguished from the other promoters of its family due to its higher conservation.
Both free-stability and base-pair stacking have been employed as good representatives of promoters, but enthalpy has not been yet. The statistical assessment has shown that the conversion of genetic information (promoters) into energy-related features indicates the binding sites of σ factors to the DNA.
Moreover, Fig. 2 was provided in order to link consensual motifs (displayed by the sequence logos) where transcription factors bind the DNA to strong signals observed in the energetic/structural features conversion. All σs matched some of their peaks to sites where RNAP binds the DNA and two families of promoters could be identified: σ54 and σ24, σ28, σ32, σ38, and σ70. The main binding sites − 10 and − 35 (σ54 extended to − 12 and − 24 instead) were observed in almost all the σs, with the exception of σ38, which did not exhibit any conserved island around − 35. This suggests that RNAP-DNA binding might be assisted by different leveling in the forces orchestrating gene transcription [9, 29, 33].
In addition, we validated the rationale found in E. coli with 15 other Gram-negative bacteria. The analysis of σ54 promoter sequences of these organisms is depicted in Fig. 3.
By analyzing the results of Fig. 3, we are able to tell apart the binding site of the σ54 transcription factor in the following organisms: A. vinelandii, B. japonicum, C. coli, K. oxytoca, K. pneumoniae, P. aeruginosa, R. eutropha, R. leguminosarum, and R. sphaeroides. The features we tested in these organisms behave in a similar way, with overlapping lines. Therefore, we suggest the analysis primarily formed upon E. coli has the potential to be further stretched to Gram-negative bacteria.
3.2 Physical parameters are employed to distinguish promoters
Structural and energetic features of the DNA have been described as discriminators of promoter sequences [9, 10, 33, 34]. To this extent, we performed the nonparametric Kruskal–Willis test in order to test the variance of averages found between promoters and control (shuffled and coding sequences). As depicted in Table 3, significant variance between the means was not found only in σ28’s enthalpy and σ54’s enthalpy and stability.
The results Table 3 conveyed suggest the features that presented significant variance between the promoters and controls (shuffled and coding sequences) might be employed in order to classify promoter sequences.
The results obtained in Table 3 have insinuated enthalpy, stability, and base-pair stacking might be good distinguishers between promoter and control (shuffled and coding) sequences. To further stretch this analysis, we opted to separately analyze each physical that protruded significant variances.
There are many features that might be employed in order to convert genetic information into, indeed, there are 125 features that have been listed [11] in this sense. Most of these properties are inclined to capture the particularities of promoter regions, which are known for being distinct from other genome locales.
Figure 4 was created in order to further explore the enthalpy variances found in Table 3 and assess if this feature enables promoters to be distinguished among control sequences. The results of Fig. 4 advocate that promoter sequences regulated by σ24, σ32, σ38, and σ70 might be classified through enthalpy, which converges to reports describing the thermostability of DNA being affected by the extra hydrogen bond found in GC-composed duplexes [26]. Apart from the chemical richness found in GC duplexes, other elements such as strand length, strand concentration, ionic strength of added salts, and water molecules were found as contributors to the melting temperature of DNA [22, 25, 32]. Hence, the several elements involved in DNA-RNAP interaction define the promoter sequences and explain the statistical differences between promoter and control (shuffled and coding) sequences from Fig. 3. DNA enthalpy has been identified as a discriminator of promoter sequences [10], being necessary to a proper gene regulation.
In the left panel, the enthalpy profiles are provided. The Y-axis indicates the enthalpy levels in kcal/mol-bp-1. The X-axis shows the nucleotide position, with the TSS located at 0. The left panel represents the boxplots of the profiles of promoter, shuffled, and coding sequences, whose Y-axis is in the same scale as the left panel
DNA free-stability is a well-documented attribute, indeed, promoter predictors succeeded in employing this feature as a form of classification [8, 27]. Figure 5 is provided in order to provide visual aid of the significant variances found in Table 3, which are found in promoter sequences associated to all σ groups with the exception of σ54. The total free-stability level of promoter sequences tends to be lower than coding regions due to the recurrent need of establishing a DNA open complex [14]. For this purpose, the hydrogen bonds between base pairs require to be broken, while an A/T duplex presents two hydrogen bonds, a G/C has three. For this process to be energetically viable, it is reasonable for the promoters to demonstrate a lower free-stability value than other genomic regions, and therefore, a higher A/T presence [8, 14]. The profiles of Fig. 5 suggest that a classifying method based on stability might succeed.
In the left panel, the free-stability profiles are provided. The Y-axis indicates the free-stability levels in kcal/mol-bp-1. The X-axis shows the nucleotide position, with the TSS located at 0. The left panel represents the boxplots of the profiles of promoter, shuffled, and coding sequences, whose Y-axis is in the same scale as the left panel
In order to differentiate promoter and non-promoter sequences, the base-pair stacking energy was tested. These interactions refer to the forces used in nucleotides connected by the phosphate group, forming the DNA backbone. In structural analysis study, the strength of this connection varies according to the composition of nucleic acids. This attribute presented a significant difference among the variances in all σ promoters (Table 3). From Fig. 6, we found that the GC binding strength corresponds to 20 piconewtons (pN); whereas AT base-pair binding strength is 14 pN; the stacking force in adjacent base pairs is estimated to be two pN [35]. We can picture base-pair stacking as the vertical relationship among the bases and DNA stability as the force in horizontal connections [14, 21]. Due to the nature of promoter sequences, they might be classified through the conserved aspect that these regulatory regions have when coded into base-pair stacking values.
In the left panel, the base-pair stacking profiles are provided. The Y-axis indicates the base-pair stacking levels in kcal/mol-bp-1. The X-axis shows the nucleotide position, with the TSS located at 0. The left panel represents the boxplots of the profiles of promoter, shuffled, and coding sequences, whose Y-axis is in the same scale as the left panel
The application of energetic/structural parameters in order to capture signals within the genome has been widely experimented [13, 24, 33]. The common rule for all these features is to be able to codify genetic information, represented by a four-letter alphabet into numeric attributes, ranging from infinity. Ryasik et al. [28] stated the importance of having genetic information represented by the use of numbers. In both eukaryotes and prokaryotes, the presence of TF-protein binding sites is a common ground, in theory. However, there are cases where the lone presence of these sets of nucleotides is not enough to capture and characterize promoter activity. In this field, Deyneko et al. [10] stated the importance of differentiating the letter conservation, i.e., presence/absence of consensual regions from a signal conservation, which encompasses structural/energetic strong signals.
4 Conclusions
The coding of genetic information into structural and physical attributes has shown capable of well-representing promoter activity. Moreover, the individual assessment of enthalpy, free-stability, and base-pair stacking proved to be a good distinguisher between promoter and control sequences. The results gathered in this study suggest that the in-silico experimentation of bacterial transcription might benefit from employing distinct ways to represent a given DNA molecule.
References
Allawi HT, Santalucia J (1997) Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry 97(36):10581–10594. https://doi.org/10.1021/bi962590c
Bansal M, Kumar A, Yella VR (2014) Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr Opin Struct Biol 25:77–85. https://doi.org/10.1016/j.sbi.2014.01.007
Barnard A, Wolfe A, Busby S (2004) Regulation at complex bacterial promoters: How bacteria use different promoter organizations to produce different regulatory outcomes. Curr Opin Microbiol 7(2):102–108. https://doi.org/10.1016/j.mib.2004.02.011
Cases I, De Lorenzo V, Ouzounis CA (2003) Transcription regulation and environmental adaptation in bacteria. Trends Microbiol 11(6):248–253. https://doi.org/10.1016/S0966-842X(03)00103-3
Chen Y, Ho JML, Shis DL, Gupta C, Long J, Wagner DS et al (2018) Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat Commun. https://doi.org/10.1038/s41467-017-02473-5
Coelho RV, de Avilae SS, Echeverrigaray S, Delamare APL (2018) Bacillus subtilis promoter sequences data set for promoter prediction in gram-positive bacteria. Data Brief. https://doi.org/10.1016/j.dib.2018.05.025
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004
De Avila e Silva S, Echeverrigaray S, Gerhardt GJL (2011) BacPP: bacterial promoter prediction-A tool for accurate sigma-factor specific assignment in enterobacteria. J Theor Biol 287:92–99. https://doi.org/10.1016/j.jtbi.2011.07.017
De Avilae SS, Forte F, Sartor TS, Andrighetti T, Gerhardt G, Longaray Delamare AP, Echeverrigaray S (2014) DNA duplex stability as discriminative characteristic for Escherichia coli σ54- and σ28-dependent promoter sequences. Biologicals 42(1):22–28. https://doi.org/10.1016/j.biologicals.2013.10.001
Deyneko IV, Kalybaeva YM, Kel AE, Blöcker H (2010) Human-chimpanzee promoter comparisons: property-conserved evolution? Genomics. https://doi.org/10.1016/j.ygeno.2010.06.003
Friedel M, Nikolajewa S, Sühnel J, Wilhelm T (2009) DiProDB: a database for dinucleotide properties. Nucleic Acids Res. https://doi.org/10.1093/nar/gkn597
Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muñiz-Rascado L, García-Sotelo JS et al (2016) RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 44(D1):D133–D143. https://doi.org/10.1093/nar/gkv1156
Ilicheva IA, Khodikov MV, Poptsova MS, Nechipurenko DY, Nechipurenko YD, Grokhovsky SL (2016) Structural features of DNA that determine RNA polymerase II core promoter. BMC Genomics. https://doi.org/10.1186/s12864-016-3292-z
Kanhere A, Bansal M (2005) A novel method for prokaryotic promoter prediction based on DNA stability. BMC Bioinformat. https://doi.org/10.1186/1471-2105-6-1
Kim D, Hong JSJ, Qiu Y, Nagarajan H, Seo JH, Cho BK et al (2012) Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002867
Krebs JE, Goldstein ES, Kilpatrick ST (2017) Lewin’s Gene XII, 12th edn. Jones & Bartlett Learning, Burlington
Kumar A, Bansal M (2017) Unveiling DNA structural features of promoters associated with various types of TSSs in prokaryotic transcriptomes and their role in gene expression. DNA Res Int J Rapid Publ Rep Genes Genomes. https://doi.org/10.1093/dnares/dsw045
Liang ZY, Lai HY, Yang H, Zhang CJ, Yang H, Wei HH, Chen XX, Zhao YW, Su ZD, Li WC, Deng EZ, Tang H, Chen W, Lin H (2017) Pro54DB: a database for experimentally verified sigma-54 promoters. Bioinformatics. https://doi.org/10.1093/bioinformatics/btw630
Lloréns-Rico V, Lluch-Senar M, Serrano L (2015) Distinguishing between productive and abortive promoters using a random forest classifier in Mycoplasma pneumoniae. Nucleic Acids Res 43(7):3442–3453. https://doi.org/10.1093/nar/gkv170
McAdams HH, Srinivasan B, Arkin AP (2004) The evolution of genetic regulatory systems in bacteria. Nat Rev Genet 5:169–178. https://doi.org/10.1038/nrg1292
Meysman P, Collado-Vides J, Morett E, Viola R, Engelen K, Laukens K (2014) Structural properties of prokaryotic promoter regions correlate with functional features. PLoS ONE 9(2):e88717. https://doi.org/10.1371/journal.pone.0088717
Morgunova E, Yin Y, Das PK, Jolma A, Zhu F, Popov A et al (2018) Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima. Elife 7:e32963. https://doi.org/10.7554/elife.32963
Ornstein RL, Rein R, Breen DL, Macelroy RD (1978) An optimized potential function for the calculation of nucleic acid interaction energies I. Base stacking. Biopolymers 17(10):2341–2360. https://doi.org/10.1002/bip.1978.360171005
Ozoline ON, Deev AA, Trifonov EN (1999) Dna bendability—a novel feature in E. coli promoter recognition. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.1999.10508295
Petruska J, Goodman MF (1995) Enthalpy-entropy compensation in DNA melting thermodynamics. J Biol Chem. https://doi.org/10.1074/jbc.270.2.746
Privalov PL, Crane-Robinson C (2018) Forces maintaining the DNA double helix and its complexes with transcription factors. Prog Biophys Mol Biol 135:30–48. https://doi.org/10.1016/j.pbiomolbio.2018.01.007
Rangannan V, Bansal M (2007) Identification and annotation of promoter regions in microbial genome sequences on the basis of DNA stability. J Biosci 32:851–862. https://doi.org/10.1007/s12038-007-0085-1
Ryasik A, Orlov M, Zykova E, Ermak T, Sorokin A (2018) Bacterial promoter prediction: selection of dynamic and static physical properties of DNA for reliable sequence classification. J Bioinform Comput Biol 16(1):1840003. https://doi.org/10.1142/S0219720018400036
SantaLucia J, Hicks D (2004) The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct 33:415–440. https://doi.org/10.1146/annurev.biophys.32.110601.141800
Singh A, Mishra A, Khosravi A, Khandelwal G, Jayaram B (2017) Physico-chemical fingerprinting of RNA genes. Nucleic Acids Res 45(7):e47. https://doi.org/10.1093/nar/gkw1236
Shahmuradov IA, Mohamad Razali R, Bougouffa S, Radovanovic A, Bajic VB (2017) bTSSfinder: a novel tool for the prediction of promoters in cyanobacteria and Escherichia coli. Bioinformatics 33(3):334–340. https://doi.org/10.1093/bioinformatics/btw629
Xu X, Zhi X, Leng F (2012) Determining DNA supercoiling enthalpy by isothermal titration calorimetry. Biochimie 94(12):2665–2672. https://doi.org/10.1016/j.biochi.2012.08.002
Yella VR, Bansal M (2017) DNA structural features of eukaryotic TATA-containing and TATA-less promoters. FEBS Open Bio 7(3):324–334. https://doi.org/10.1002/2211-5463.12166
Yella VR, Kumar A, Bansal M (2018) Identification of putative promoters in 48 eukaryotic genomes on the basis of DNA free energy. Sci Rep. https://doi.org/10.1038/s41598-018-22129-8
Zhang TB, Zhang CL, Dong ZL, Guan YF (2015) Determination of base binding strength and base stacking interaction of DNA duplex using atomic force microscope. Sci Rep. https://doi.org/10.1038/srep09143
Funding
This work was supported by Universidade de Caxias do Sul and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Martinez, G.S., de Ávila e Silva, S., Kumar, A. et al. DNA structural and physical properties reveal peculiarities in promoter sequences of the bacterium Escherichia coli K-12. SN Appl. Sci. 3, 740 (2021). https://doi.org/10.1007/s42452-021-04713-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-021-04713-2